Genotype-Phenotype maps of ancestral proteins
A long-standing question
This is currently my Ph.D. project in Joe Thornton's lab at the University of Chicago.
Why some phenotypes evolve whereas other alternative ones have never been observed? This question has puzzled naturalists, paleontologists and developmental biologists for over a century. Even R. Lewontin dedicated an essay to this very question:
The real problem for the evolutionist is not to explain the kinds of organisms that have actually ever existed. The real problem for the evolutionist is how it is that most kinds of potential and seemingly reasonable organisms have never existed.
-- Richard Lewontin, Four Complications in Understanding the Evolutionary Process, 2003

One reason may be that the forms we observe today are the most optimal forms that can be produced; an alternative reason, however, is that these forms are the most likely to arise by mutation from an ancestral point.
These two hypotheses are, as almost everything in biology, two extremes of a continuum. In reality, both processes are likely to interact in complex ways creating the amazing diversity of forms that we observe today. However, disentangling the relative roles and interactions of these kinds of causes is easier said than done. For most biological systems, this proves to be intractably complex.
A diverse world of proteins
When thinking about diversity of forms, proteins are unlikely to be the first thought. However, proteins have an incredible diversity of structures and functions. What's more, combining evolutionary genetics and protein biochemistry can open the door to understanding in detail how and why did proteins evolve their present day phenotypes.
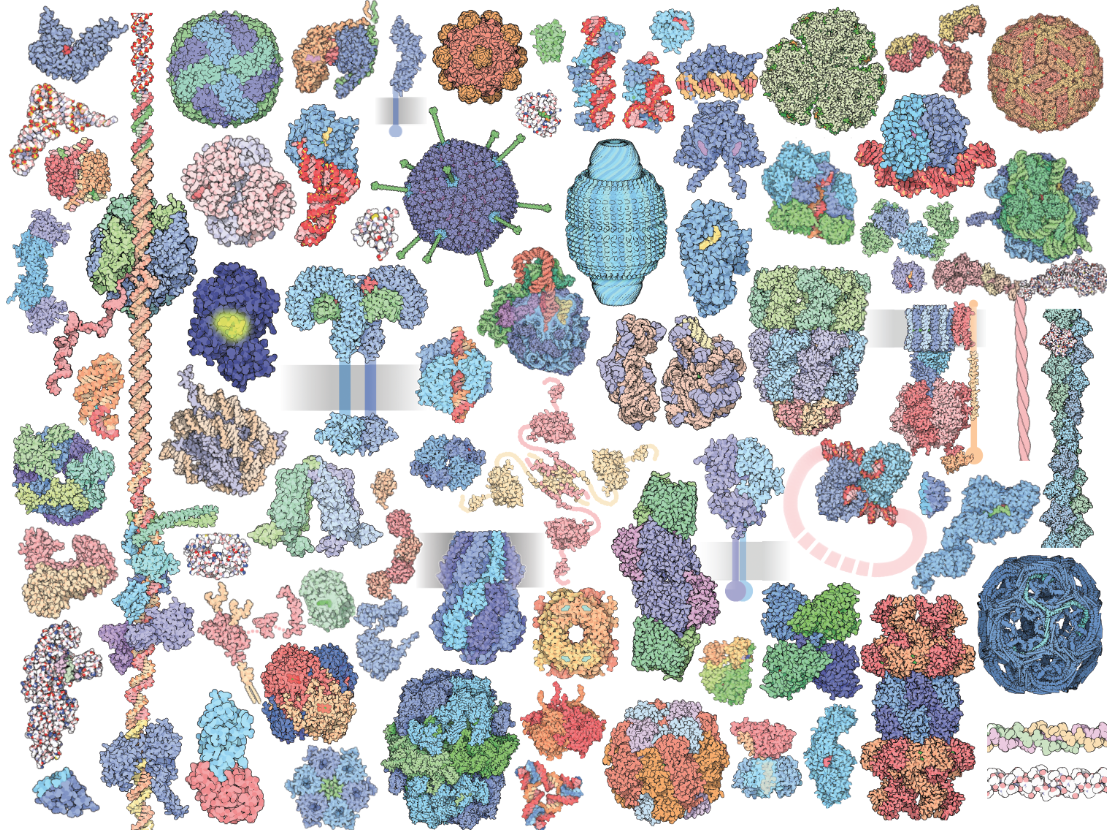
Experimental manipulation on proteins allows one to establish clear connections between sequence-structure-function, making them ideal systems to systematically explore the effects of mutations on the production of phenotypic diversity.
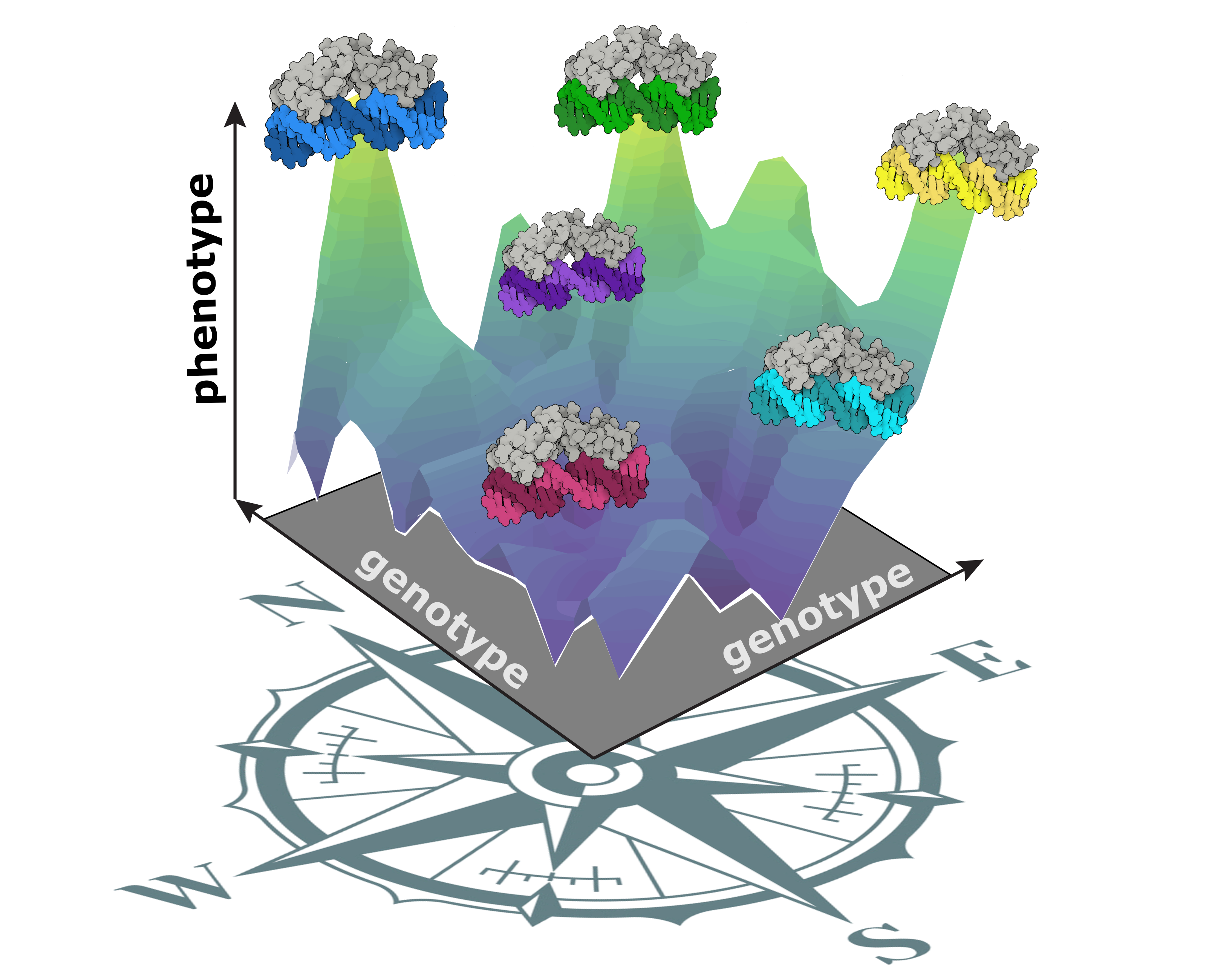
Towards a complete GP-map
Since evolution is a historical process, explaining present-day phenotypic variation requires knowing the range of phenotypic variation that what was possible at some relevant point in the past.
I'm combining ancestral sequence reconstruction and large-scale mutagenesis to experimentally reconstruct the genotype-phenotype landscape of ancestral proteins. I'm applying this approach to a molecular system essential to vertebrate development and physiology: the regulatory module consisting of steroid hormone receptors (SRs) and their DNA-binding elements.
These experiments will allow me to characterize all the forms of variation in DNA-binding function that could possibly been produced at relevant points during the evolutionary history of SRs and all the mutational trajectories to each of the new phenotypes.